Researchers Propose Novel Mechanism for the Treatment of Friedreich’s Ataxia
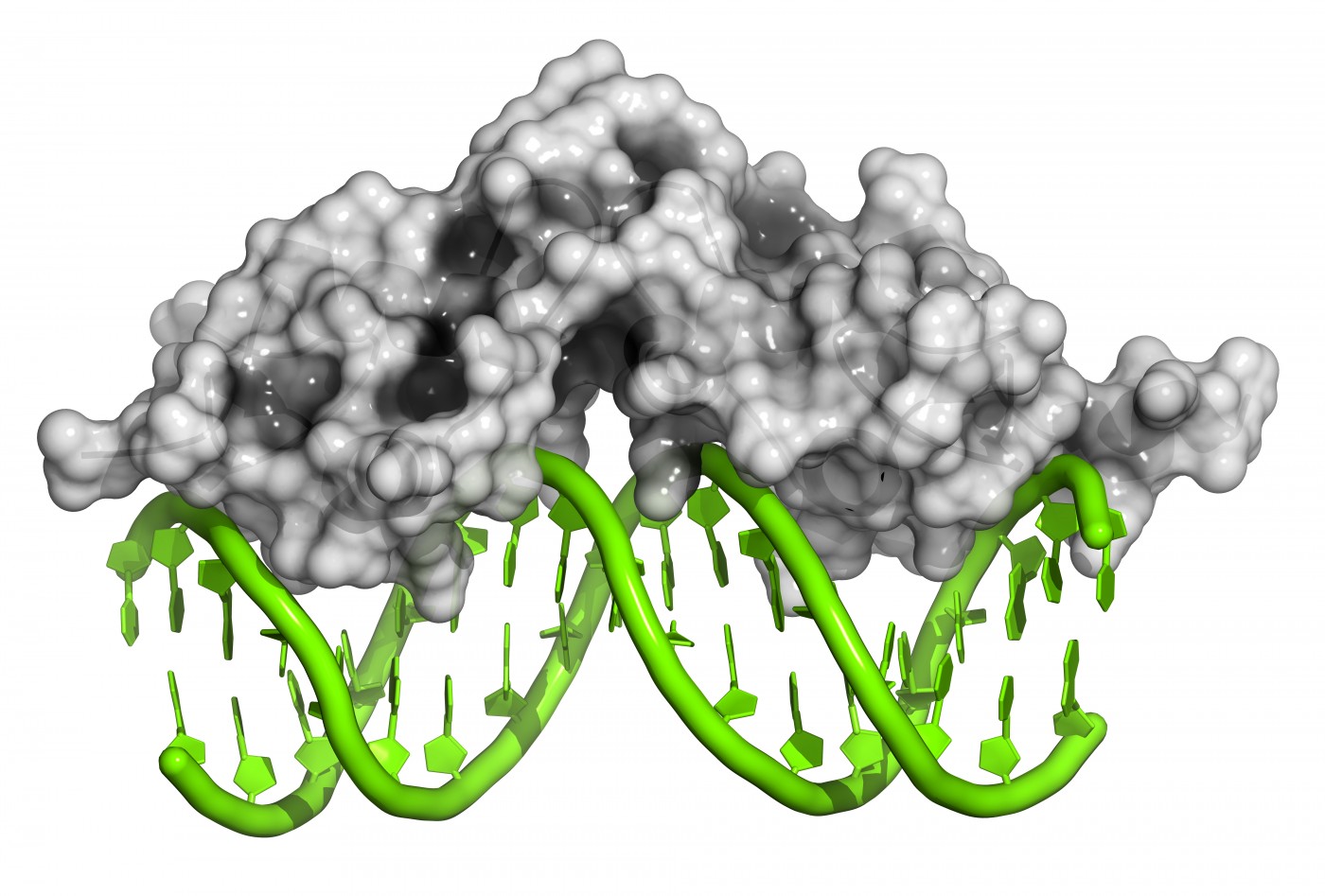
It is well known that a DNA mutation causes Friedreich’s ataxia by reducing the production of frataxin protein. Patients with Friedreich’s ataxia often have several hundred “GAA repeats” in their DNA, whereas unaffected individuals have no more than 40 GAA repeats. What is less known is how exactly GAA repeats interfere with the process of frataxin gene transcription into frataxin mRNA and subsequent translation into frataxin protein.
Dr. Jill Sergesketter Butler and Dr. Marek Napierala at the Stem Cell Institute inside the University of Alabama at Birmingham are working to unravel the mechanisms at play. In a recent article published in the journal Transcription, entitled “Friedreich’s Ataxia – A Case of Aberrant Transcription Termination?” the team identified how protein translation may be prematurely truncated due to R-loop structures, antisense transcription, and heterochromatin formation. Their article explained this model in greater detail.
“The critical link between the DNA mutation and silencing of FXN is missing,” wrote the authors, to support their investigation into the field. Previous studies showed that expanded GAA repeats do not interfere at the level of transcript stability, pre-mRNA gene splicing, or RNA foci. However, there is a notable body of evidence showing that histone protein modifications to DNA play a key role in decreasing frataxin protein expression. Treating patients with histone deacetylase (HDAC) inhibitors seems to help, but histone methyltransferase inhibitors do not. Before combining medications is pursued as a treatment option for patients, more work is needed.
To answer the unmet need, groups have explored R-loop structures. R-loop structures seem to lead to antisense transcription in cells, then activation of silencing RNA pathways. This leads to local changes in chromatin and early truncation of frataxin gene transcription. “This model, if validated in Friedreich’s ataxia cells, could shift therapeutic efforts toward alleviating R-loop formation at the GAA repeats rather than targeting the silencing histone marks that appear to be a consequence of the DNA-RNA hybrid formation,” stated Dr. Butler and Dr. Napierala. “Therefore, targeting pathogenic R-loops has to be conducted very precisely and specifically without affecting physiologically important DNA-RNA hybrids.”
By understanding pathologies in Friedreich’s ataxia such as R-loop structures, scientists can develop therapies that treat causes rather than symptoms of disease, something that is lacking for patients with Friedreich’s ataxia. As scientists learn more and more about R-loops, they are discovering they are less rare than previously thought, indicating that this is an important avenue to consider when targeting specific causes of disease.